A subscription to JoVE is required to view this content. Sign in or start your free trial.
Method Article
Improvement of Bacillus subtilis Spore Enumeration and Label Analysis in Flow Cytometry
In This Article
Summary
This protocol focuses on the use of flow cytometry and counting beads to quantify bacterial spores labeled with ethidium bromide. The method is also efficient for analyzing the covalent coupling of proteins on the surface of intact spores.
Abstract
The spores of Bacillus subtilis have already been proposed for different biotechnological and immunological applications; however, there is an increasing need for the development of methodologies that improve the detection of antigens immobilized on the surface of spores together with their quantification. Flow cytometry-based analyses have been previously proposed as fast, reliable, and specific approaches for detecting labeled cells of B. subtilis. Herein, we propose the use of flow cytometry to evaluate the display efficiency of a fluorescent antibody (FA) on the surface of the spore and quantify the number of spores using counting beads.
For this, we used ethidium bromide as a DNA marker and an allophycocyanin (APC)-labeled antibody, which was coupled to the spores, as a surface marker. The quantification of spores was performed using counting beads since this technique demonstrates high accuracy in the detection of cells. The labeled spores were analyzed using a flow cytometer, which confirmed the coupling. As a result, it was demonstrated that DNA labeling improved the accuracy of quantification by flow cytometry, for the detection of germinated spores. It was observed that ethidium bromide was not able to label dormant spores; however, this technique provides a more precise determination of the number of spores with fluorescent protein coupled to their surface, thus helping in the development of studies that focus on the use of spores as a biotechnological platform in different applications.
Introduction
Bacillus subtilis is a rod-shaped, gram-positive bacterium that is able to produce quiescent spores when environmental conditions do not allow cell growth1. Spores are extremely stable cell forms and those of several species, including B. subtilis, are widely used as probiotics for human and animal use2. Due to its resistance and safety properties, the spore of B. subtilis, which displays heterologous proteins, has been proposed as a mucosal adjuvant, a vaccine delivery system, and an enzyme-immobilization platform3,4.
To obtain spores from B. subtilis, it is necessary to expose it to nutrient deprivation using a special culture medium. After obtaining and purifying these spores, one must quantify them to improve test efficiency5,6. Thus, certain methods are applied to analyze the concentration of the spores obtained. Plate counting and a Petroff-Hausser chamber, also known as a counting chamber, can be used. The latter was originally developed to determine the concentration of blood cells; however, it is possible to use it in the field of microbiology for spore counting7,8. Despite being the standard method used for cell counting, reading is laborious since this method is completely manual and its accuracy depends on the experience of the operator.
Flow cytometry-based (FC) analyses have been previously proposed as fast, reliable, and specific approaches for detecting labeled cells of Bacillus spp. The use of flow cytometry counting beads has guaranteed reproducibility in cell counting in routine examinations (absolute count of CD4 and CD8 T lymphocytes) and in the development of research involving particles capable of being detected and counted using flow cytometry9. Godjafrey and Alsharif suggested the use of counting beads for FC quantification of unlabeled spores10. The use of flow cytometry was described for the monitoring of sporulation in Bacillus spp. via labeling the spore DNA10,11,12,13. Yet another study used FC to evaluate the amount of fluorescently labeled proteins on the spore surface15.
This study sought to use commercial counting beads to ensure a standard of reproducibility with respect to event counting using flow cytometry. Herein, we suggest the use of counting beads for cell counting in FC to refine spore enumeration and evaluate the coupling efficiency of fluorescently labeled antibodies on the spore surface.
Protocol
See the Table of Materials for details related to all materials, instruments, and software used in this protocol.
1. Flow cytometry setting
- Alignment of optical parameters of flow cytometer coupled to a computer
- Log in to Cytometer software.
- From the software workspace, select Cytometer | Startup and wait a few minutes | Clean mode | Sit fluids.
NOTE: Air bubbles and obstructions were removed during the cytometer initiation process, prior to sample acquisition.
- Quality control
- Use the quality control reagent to adjust the voltages of the photomultiplier tubes and evaluate their sensitivity.
- Prepare the quality control reagent in a polystyrene tube.
- For defining the baseline, prepare the suspension beads by adding 0.5 mL of diluent (10 mM filtered PBS, pH 7) and 3 drops of beads to the labeled tube.
- Mix the bead vial by gentle inversion.
- From the software workspace, select Cytometer | CST to connect to the CST interface. Wait a few minutes and observe the message cytometer disconnected.
- Attach the polystyrene tube containing the quality control reagent to the flow cytometer probe.
- Use the quality control reagent to adjust the voltages of the photomultiplier tubes and evaluate their sensitivity.
- Verification of the configuration of the cytometer
- In the System Summary window, check that the cytometer configuration is appropriate for the experiment.
- Select the setup beads batch ID corresponding to verify that the setup beads lot ID selected matches the current lot of CS&T research beads.
- Performance check
- Select the option Check Performance and click Run.
- Upon completion of the performance check, the performance results will be displayed. View the Cytometer Performance Report. Either click Finish to complete the performance check or remove the tube from the cytometer and review the results in the System Summary window.
- Observe the final result of the morphometric and fluorescence sensitivity analysis that will appear in the | system summary| cytometer performance result| with the status: PASSED
2. Preparation of the spores
- After sporulation, rinse the spores 3x with ultrapure ice-cold water by centrifugation at 17,949 × g for 10 min.
- Resuspend the pellet obtained from the last centrifugation with 10 mL of ultrapure water.
- Autoclave the spores for 45 min at 121 ºC for inactivation of the vegetative cells.
NOTE: Previous tests carried out by our group comparing different inactivation methods demonstrated that the conditions mentioned in step 2.3 presented high efficiency, showing no colony formation in the plating analysis method on the LB agar medium.
3. Quantification of autoclaved spores using flow cytometry
- Take 50 µL of autoclaved spores and incubate them protected from light with ethidium bromide (EtBr, 10 mg/mL diluted in water) at a dilution factor of 0.05% v/v for 30 min (Figure 1A).
- Wash the spores 3x with 1x phosphate-buffered saline (PBS) by centrifugation at 17,949 × g for 10 min and resuspend in 1x PBS.
- Add 10 µL of beads, following the manufacturer's recommendations for dilution.
- Analyze the sample using a flow cytometer as described in step 4.
NOTE: It is important to emphasize that the beads should be pipetted as homogeneously as possible since disparities were observed in the calculations when this step was not well executed.
4. Analysis using flow cytometry
- Log in to Cytometer software.
- From the software workspace, select Cytometer | Clean mode | Sit fluids.
- Adjust to the workspace to determine the analysis in flow cytometry.
- Define the gates for the analyses (Figure 2).
- Define the gating strategy based on the morphometric and fluorescence characteristics of the particles based on the negative control (unmarked spores).
- To determine cell morphometry, choose Dotplot graphic for analyses of FSC-A parameters on the x-axis and SSC-A on the y-axis.
- To determine fluorescence, choose FL3 Dotplot plots on the y-axis using FL5 on the x-axis and create the gates in four quadrants.
- Define the gates for the analyses (Figure 2).
- Use a 12 x 75 mm stoppered polystyrene tube containing unlabeled samples, previously labeled with single-color EtBr, single-color APC, and multicolor EtBr+ APC.
- Mix the tube very gently containing the negative control and attach it to the flow cytometer probe.
- Click on Acquire.
- Set the power of the lasers by navigating to Cytometer | Parameters | FSC (375) and SSC (275).
- Set the threshold to (500) Cytometer | Threshold.
- To remove autofluorescence, analyze the dye-free sample containing only spores.
- Adjust the voltages for the filter detectors: FL3 (603) and FL5 (538) to discriminate negative and positive populations with respect to the chosen fluorescence in the controls. Cytometer | Parameters.
- Cytometer | Compensation | Set FL5/FL3 setup offset to 1.0.
- After morphometry and fluorescent analysis, configure the device for Acquisition 30,000 events | Experiment | Experiment layout | Acquisition |30,000 events….
- After adjusting the parameters, acquire data for the samples that are labeled and contain beads in the flow cytometer.
- Calculate spore concentration as described by the manufacturer's instructions using equation (1).
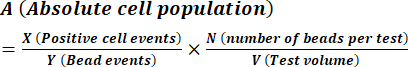 (1)
(1)
NOTE: For this study, the bead quantification method was compared to the Petroff-Hausser counting chamber analysis. In addition, the labeling of autoclaved spores was compared to the labeling of non-autoclaved spores.
5. Estimation of the protein coupling index on a spore surface using flow cytometry
- Harvest by centrifugation of 50 µL of spores (103/µL) 17,949 × g for 10 min and then, resuspend with 25 µL of 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide (EDC) (300 mM)).
- Incubate for 15 min at room temperature.
- Add 25 µL of N-hydroxysulfosuccinimide (NHS) (50 mM) to the spore suspension and incubate it at room temperature for 30 min.
- Wash the spores 3x with 1x PBS by centrifugation as described in step 5.1.
- Add fluorescent protein and leave the samples overnight at 15 °C.
NOTE: The fluorescent protein used in this work was an APC-labeled anti-human IL-10 antibody. This antibody was used as a model for study, though the application of other fluorescent molecules is possible since EDC/NHS promote a covalent ligation between -COOH and -NH2 groups present in proteins on the spore surface. - Wash the spores as per step 5.3.
- Add 10 mg/mL of ethidium bromide diluted at 1:50, then leave for 1 h on ice and protected from light (Figure 1B).
- Wash the spores as per step 5.3.
- Repeat step 4.
- To determine fluorescence, change the parameters FL3 on the X axis and FL5 on the Y axis of the dotplot.
- Analyze the sample using a flow cytometer with the blue laser (488 nm) at FL3 (670 LP filter) and the red laser (633 nm) in FL5 (660/20 filter), as described in step 4.
NOTE: The same samples were analyzed for immunofluorescence on slides under a microscope, at a magnification range of 1,000 x, and using the following filters: FITC 480/30 nm (green) and TRITC 540/25 nm (red).
Results
In autoclaved spore (AS) samples, 2 × 103 spores/µl and 1 × 103 spores/µl were detected by using counting beads and the Petroff-Hausser method, respectively (Figure 2).
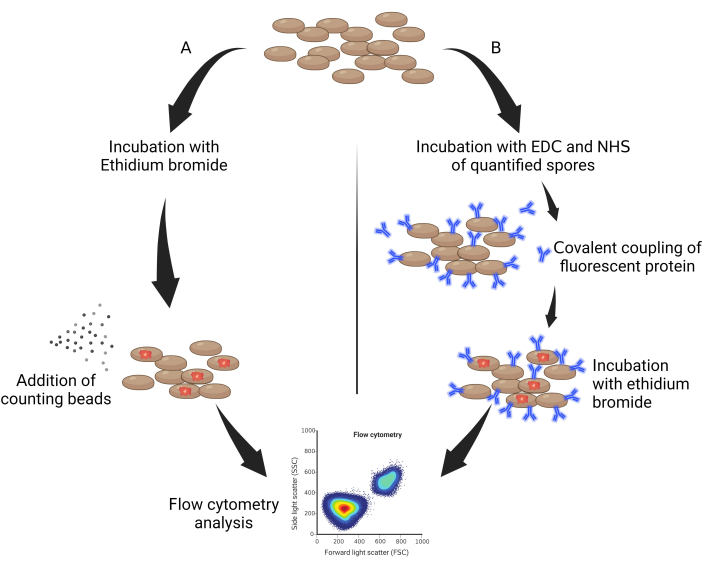
Figure 1: General scheme of quantification of spores. (A) Spores labeled with...
Discussion
Traditional methods, such as plate counting of colonies, are not only time-consuming, but also need viable cells and do not allow for quantification of inactivated spores5. The Petroff-Hausser chamber is an alternative methodology, but it requires an experienced microscopist to perform it. Flow cytometry has proven to be a useful alternative for this purpose.
Genovese et al.12 described the use of flow cytometry for the quantification of viable c...
Disclosures
The authors have no conflicts of interest to disclose.
Acknowledgements
This study was financed in part by the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior-Brasil (CAPES)-Finance Code 001; Governo do Estado do Amazonas with resources from Fundação de Amparo à Pesquisa do Estado do Amazonas-FAPEAM; Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq). The authors thank the Program for Technological Development in Tools for Health PDTIS-FIOCRUZ for use of its facilities.
Materials
| Name | Company | Catalog Number | Comments |
| (N-hydroxysuccinimide) (NHS) | Sigma | 130672 | |
| Anti-human fluorescent antibody | BioLegend | 501410 | APC anti-human IL-10 |
| Anti-mouse fluorescent antibody | Thermo Scientific | A32723 | Alexa Fluor Plus 488 |
| BD FACSCanto II | BD | Flow cytometer | |
| BD FACSDiva Cytometer Setup & Tracking Beads Kit (use with BD FACSDiva software v 6.x) | BD | 642412 | Quality control reagent |
| BD FACSDiva Software v. 6.1.3 | BD | 643629 | Software |
| Centrifuge MegaFuge 8R | Thermo Scientific | 75007213 | |
| Counting Beads | BD | 340334 | TruCount Tubes |
| Eclipse 80i | Nikon | Fluorescent Microsope | |
| Ethidium Bromide | Ludwig Biotec | ||
| Phosphate buffered saline | Sigma-Aldrich | A4503 | |
| Plastic Microtubes | Eppendorf | ||
| Polystyrene tube | Falcon | 352008 | 5 mL polystyrene tube, 12 x 75 mm, without lid, non-sterile |
References
- McKenney, P. T., Driks, A., Eichenberger, P. The Bacillus subtilis endospore: assembly and functions of the multilayered coat. Nature Reviews. Microbiology. 11 (1), 33-44 (2013).
- Cutting, S. M. Bacillus probiotics. Food Microbiology. 28 (2), 214-220 (2011).
- Ricca, E., Baccigalupi, L., Cangiano, G., De Felice, M., Isticato, R. Mucosal vaccine delivery by non-recombinant spores of Bacillus subtilis. Microbial Cell Factories. 13, 115 (2014).
- Falahati-Pour, S. K., Lotfi, A. S., Ahmadian, G., Baghizadeh, A. Covalent immobilization of recombinant organophosphorus hydrolase on spores of Bacillus subtilis. Journal of Applied Microbiology. 118 (4), 976-988 (2015).
- Harrold, Z., Hertel, M., Gorman-Lewis, D. Optimizing Bacillus subtilis spore isolation and quantifying spore harvest purity. Journal of Microbiological Methods. 87 (3), 325-329 (2011).
- Nicholson, W. L., Setlow, P. Sporulation, germination and outgrowth. Molecular biological methods for Bacillus. , (1990).
- Mora-Uribe, P., et al. Characterization of the adherence of Clostridium difficile spores: the integrity of the outermost layer affects adherence properties of spores of the epidemic strain R20291 to components of the intestinal mucosa. Frontiers in Cellular and Infection Microbiology. 6, 99 (2016).
- Paidhungat, M., Setlow, P. Role of ger proteins in nutrient and nonnutrient triggering of spore germination in Bacillus subtilis. Journal of Bacteriology. 182 (9), 2513-2519 (2000).
- Schnizlein-Bick, C., Spritzler, J., Wilkening, C., Nicholson, J., O'Gorman, M. Evaluation of TruCount absolute-count tubes for determining CD4 and CD8 cell numbers in human immunodeficiency virus-positive adults. Site Investigators and The NIAID DAIDS New Technologies Evaluation Group. Clinical and Diagnostic Laboratory Immunology. 7 (3), 336-343 (2000).
- Godfrey, A., Alsharif, R. Rapid enumeration of viable spores by flow cytometry. US Patent. , (2003).
- Karava, M., Bracharz, F., Kabisch, J. Quantification and isolation of Bacillus subtilis spores using cell sorting and automated gating. PLoS ONE. 14 (7), e021989 (2019).
- Genovese, M., Poulain, E., Doppler, F., Toussaint, R., Boyer, M. Bacillus spore enumeration using flow cytometry: A proof of concept for probiotic application. Journal of Microbiological Methods. 190, 106336 (2021).
- Trunet, C., Ngo, H., Coroller, L. Quantifying permeabilization and activity recovery of Bacillus spores in adverse conditions for growth. Food Microbiology. 81, 115-120 (2019).
- Tehri, N., Kumar, N., Raghu, H., Vashishth, A. Biomarkers of bacterial spore germination. Annals of Microbiology. 68, 513-523 (2018).
- Isticato, R., Ricca, E., Baccigalupi, L. Spore adsorption as a nonrecombinant display system for enzymes and antigens. Journal of Visualized Experiments. 145, e59102 (2019).
- Song, M., et al. Killed Bacillus subtilis spores as a mucosal adjuvant for an H5N1 vaccine. Vaccine. 30 (22), 3266-3277 (2012).
Reprints and Permissions
Request permission to reuse the text or figures of this JoVE article
Request PermissionThis article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. All rights reserved